Introduction
The classic method to generate antibodies for research purposes is to immunize an animal with the protein of interest (or a part thereof), and allowing the animal to elicit an immune response to generate antibodies that specifically bind to the immunised protein. These antibodies can be purified from the serum or plasma. Standard protocols for this process exist for many decades. Because of their high specificity, such immune reagents are generally preferred over non-immune reagents.
However, antibodies generated this way have not always been used successfully or were not robust enough (1, 2). And the involvement of animals means variations in antibody performance from one animal to the next (3). Certain types of molecules, and even certain proteins, are not fit to elicit an immune response, and therefore this classic way of generating specific reagents is not always feasible. Hence the interest for synthetic (immuno-like) reagents.
When antibodies are generated and produced without the involvement of animals, they are regarded as synthetic antibodies, or recombinant antibodies (4). In addition, non-antibody based alternatives have emerged as well, such as several forms of scaffold proteins displaying diversified amino acid sequences with specific affinity binding properties, chemically stabilised nucleic acid oligomers called aptamers, and molecularly imprinted polymers (MIPs). They all have in common that the binding site is optimized for highly specific and strong interaction with a particular part of the target molecule, called the epitope. This 2-part series will discuss these different groups of reagents to depth.
This is part 2 on the synthetic immune reagents as alternatives to antibodies raised in animals. In this part we focus on scaffold proteins and on the aptamers. In part 1, we have focused on the recombinant antibodies and on Molecularly Imprinted Polymers (MIPs built around the analyte of choice).
Scaffold proteins
What are they?
Affinity binders based on scaffold proteins have been designed and used as alternatives to antibodies (5), including Abdurins, Affibodies, Affimers, Affitins, Anticalins, Bicyclic peptides, DARPins, Fynomers, Kunitz domains, and Monobodies. They all follow the same concept: a stable scaffold protein is used to display diversified amino acid sequences at exposed surface sites. The best affinity binders are selected using an appropriate display platform (phage, yeast or ribosome display) in the same way as for the selection of recombinant antibodies (see part 1). From this follows that also for this type of affinity binders, the quality of the screen depends on the quality of the library, and on the quality of the material to screen against. And, as discussed for recombinant antibodies, also a counter screen to address potential cross-reactivity should be involved.
The diverse range of non-antibody protein scaffolds provides opportunity to exploit native characteristics of the relevant target-binding domain and to produce tailored applications (5). The advantage of a tailored approach is that wildtype functions of each binding domain can be leveraged to overcome limitations of the traditional immunoglobulin antigen binding site, such as the modular construction of DARPins (6), convex binding surfaces of Affibodies (7), protease targeting in Kunitz domains (8), or the versatility of the FN3 domains in monomers to design context-sensitive drugs or biosensors (9).
Because of the small size and the low-cost of production, non-immunoglobulin scaffolds as alternatives to antibodies are being evaluated in clinical trials (10) and they are likely to enter the commercial tool affinity binder market some time in the future. As with antibodies, validation for specific purposes and batch quality control remain necessary.
Examples
Monobodies are based on a human Fibronectin (adhesive glycoprotein) scaffold: The type III domains (FN3) are folded in a similar way as the antigen binding site of an antibody (Fig1). After the tenth FN3 domain was validated as a scaffold for molecular recognition (11, 12), the loops in the domain were engineered to different lengths and in different amino acid diversities to seek high specific binding characteristics to desired targets. Although a serine/tyrosine binary code is sufficient to generate nanomolar affinity interactions in an antigen-binding fragment (13, 14), subsequent studies revealed that the full amino acid diversity is superior over the binary code to find picomolar to low nanomolar binders (15).
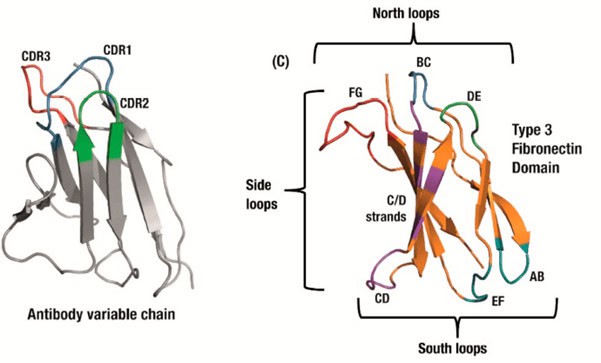
Figure 1 (copy of Fig 1B in Chandler and Buckle 2020, ref 18). Antibody domains use a set of three hypervariable binding loops (CDRs) to form a complementary region to a target binding site (left). Fibronectin type III (FN3) domains have a comparable set of analogous loops which can be engineered for similar binding function, as well as an expanded binding footprint in the side and ‘south’ loops (right).
Because monobodies have no cystein residues, they can readily be expressed in intracellular compartments with a reducing environment without losing integrity. In addition, the capability of monobodies as crystallization chaperones facilitates studies on protein structure, which in turn advances the understanding of biological processes and improved structure-guided design (16). When monomers are designed for therapeutic targets, they may be called AdnectinsTM (17). Other monomer variants used for potential therapeutics include intrabodies, E10-FN3, Centyrins, FN3Con and Tenascin-FN3 (18).
The Abdurin scaffold was developed to address the disadvantage of the shorter half-life for the smaller antibody-like scaffolds for in vivo applications. Abdurins are derived from the CH2 domain of human IgG. They are small (12.5kDa) and they are stable as monomers in the absence of glycosylation and of other IgG subunits (19). Its structure strongly resembles the antigen binding site of IgG (fig 2), thus its loops can be engineered for the generation of large libraries of binders to target molecules of interest. High affinity binders to human EphA2 have been generated and used to specifically target tumours in a xenograft model (20). However, initial attempts to use abdurins as drug conjugates for tumour therapy were not as successful as one would have hoped for (21).
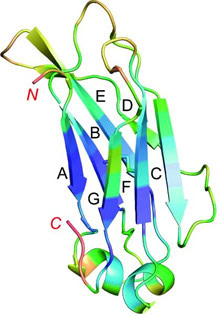
Figure 2. Structure of the CH2 antibody domain (copy of figure 1 of Prabakaran et al 2008, ref 19).
Affibodies are derived from the three helical Z domain of Staphylococcus Protein A, which is involved in the binding of the IgG Fc domain (22), and they were originally designed by randomization of 13 solvent-accessible surface residues on two of the three helices within this Z domain (Fig 3) for the identification of specificity to Taq DNA polymerase, human insulin and ApoA1 (23). The Affibody molecule is very robust, and has generated useful biotechnological tools, such as the alkali-resistant active component in a kit for affinity purification of mAbs (MabSelect SuRe) or as part of a hot start PCR kit, where the Affibody molecule improves the fidelity of the DNA polymerase and by making the polymerase survive a larger number of heat cycles (Phusion Hot Start II DNA Polymerase) (24).
Radiolabeled affibody molecules can provide highly specific and sensitive imaging. A phase I/II clinical trial showed that 68Ga-labeled affibody molecules permit an accurate and specific measurement of HER2 expression in breast cancer metastases (25). Radiolabelled affibodies specific to EGFR improve imaging of EGFR-expressing tumours (26).
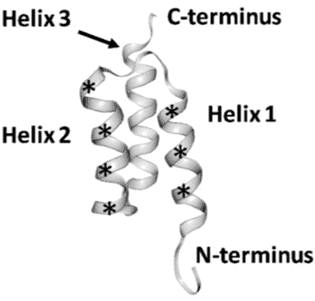
Figure 3. Structure of an affibody scaffold. Asterisks mark the segments where randomized amino acids are located. (Copy of Figure1 in Tolmachev and Orlova 2020, ref 25).
More than six different Affibody molecules have been combinatorially fused with antibodies to form functional multispecific proteins called AffiMabs (27, 28) with superior in vivo therapeutic activity in a xenograft tumor model when administering a bispecific molecule based on the EGFR-specific antibody cetuximab and a HER3-specific Affibody molecule (27). And engineered multivalent affibodies to HER3 enhanced the downregulation in multiple cancer cell types (29). A HER2-specific affibody was conjugated to a near-infrared silicon phthalocyanine dye, IRdye700DX (IR700), and applied to a xenograft HER2 cancer (BT474) in mice. Light-irradiation activates IR700 to kill the cells at the conjugate’s location, thus showing efficacy for this type of potential therapy (30). Affibodies against Epstein Bar virus (EBV) protein LMP2A were able to reduce viability of EBV-positive cell lines and of nasopharyngeal carcinoma (NPC)-derived cell lines (31).
The adhiron scaffold is a synthetic protein originally based on a phytocystatin consensus sequence. Adhirons display remarkable thermal stability (Tm = 101°C) (32). The adhiron scaffold is related in structure to a previously reported scaffold engineered from human stefin A (33). Binding proteins derived from these two scaffolds are now referred to collectively as Affimer proteins, or Affimers.
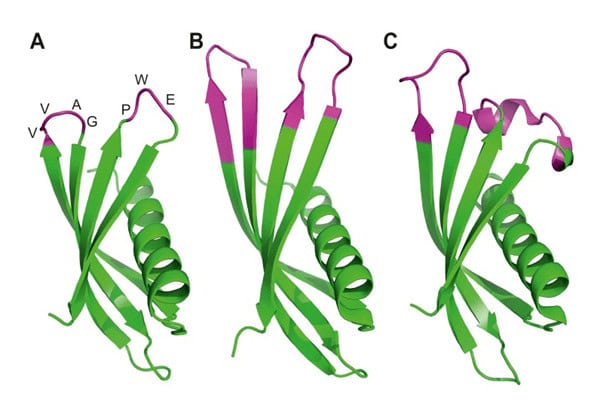
Figure 4. Ribbon diagrams of the crystal structures for Affimer (Adhiron) reagents. The unchanged loops in pink (A), specific to p300 (B) and to SUMO protein (C) with the variable regions in pink. (Copied from Tiede et al, 2017, ref 34).
Affimers have demonstrated to be good alternatives to antibodies as research reagents as demonstrated by the generation and successful use of affimers against 12 different targets, including a small organic molecule (34), and a recent report concluded that for both IL37B and proinsulin, affimers show higher specificity in purifying their target proteins from human plasma compared to monoclonal antibodies (35). They also appear superior for quantification in LC/MS (Liquid Chromatography before Mass Spectrometry) as demonstrated for serum RAGE when they detect 25% lower sRAGE levels than the validated antibody-based method (36). And they are an excellent tool as anti-idiotypic reagents for pharmacokinetic studies of therapeutic antibodies (37). Finally, the manufacturer of the affimers, Avacta, has developed a rapid test to SARS-CoV-2.
The affitin scaffold is commonly derived from Sac7d, a dsDNA-binding protein from the hyperthermophilic archaeon Sulfolobus acidocaldarius. Sac7d is chemically and thermally stable and is resistant to extreme pH. Its molecular organization is quite simple, being small (66 residues) and monomeric, lacking a disulfide bridge, and possessing only one structural domain (Figure 5). The residues involved in DNA binding can be varied to make the scaffold bind other targets of interest (38). After showing that affitins are fit as a scaffold alternative to antibodies by selection of specific reagents through ribosome display librabries (39), and fit to use for affinity purification (40), they have been successfully used in diagnostics to identify bacterial species (41). Sac7d-based Affitins are good for detection-, capture-, and inhibition-based applications, such as one-step ELISA and Western blots, immunolocalization, biosensors, protein-chip array, affinity chromatography, magnetic fishing, enzymatic and intracellular inhibition, and imaging (42).
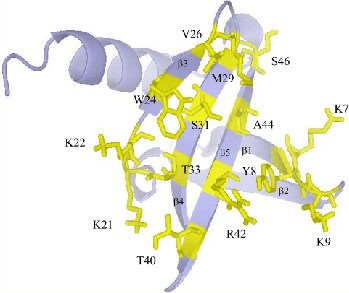
Figure 5. Schematic representation of wild-type Sac7d. Residues involved in DNA binding and that were substituted are shown in yellow. (copied Figure 1 from Mouratou et al, 2007, ref PMID: 17984049
In addition to Sac7D, affinitins can also be based on similar archaea proteins such as 63 residues Sso7D (43) and the 60 residues Aho7C (42), all benefitting from high thermostability and small size without cysteins (no sulphur bridges for secondary structures).
Anticalins are derived from proteins of the lipocalin family, a family of diverse proteins that normally serve for the storage or transport of hydrophobic small molecules, such as biliverdin IXγ, a butterfly chromophore, retinal, an important pigment for visualization, or pheromones. Despite low mutual sequence homology, lipocalins share a conserved barrel of eight antiparallel β-strands as their central folding motif. At one end of this structure, four loops connect each pair of β-strands and these loop regions are involved in the specific binding of their target molecule. Thus, although being composed of a single polypeptide chain and being much smaller, the lipocalins resemble immunoglobulins, owing their vast potential of antigen specificities to the six hypervariable loops on top of a rigid framework. Initial anticalins were developed from the Pieris brassicae (butterfly large white) anticalin BBP (Bilin-Binding Protein) by panning a phage library for specificity to fluorescein (44) and to digoxigenin (45).
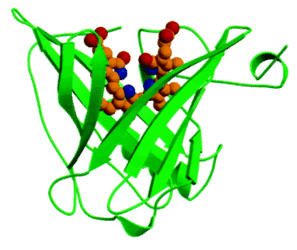
Figure 6. The lipocalin domain of BBP bound to the butterfly chromophore biliverdin IXγ (PMID: 10051566).
Since then, anticalins have been developed specific to small molecules, peptides and proteins based on the human Lipocalin 2 (Lcn2/NGAL) scaffold (46 and references therein, 47) and on human Lipocalin 1 (Lcn1/TLC) scaffold (48). Because of the small size, stability and capacity to specifically bind small molecules, like hapten, these human scaffolds are excellent vectors for pharmaceutical interventions. Hence anticalins are investigated in preclinical and clinical stages for therapy against a variety of diseases (49).
The butterfly BBP scaffold remains useful as a detector for non-human targets, example for herbicide quantification (50).
Natural bicyclic peptides are generated as a protective toxin by multiple organisms. Examples are moroidin produced by the seed of Celosia argentea (plumed cock’s comb) that inhibits eukaryotic cell division by interference in tubulin polymerization (51), and phalloidin and a-amanitin produced by mushrooms interfering in the depolymerization of actin filaments (52), and inhibiting RNA polymerase in rats, respectively (53).
For the development of new specific binders, either synthetic libraries are constructed, or libraries are generated through biological production by ribosomal synthesis, phage-based production or through the mRNA display method (54). Thus, these small and stable peptide-based reagents may serve as therapeutics, such as enzyme inhibitors (55, 56, 57), inhibitors of protein interactions (58, 59), receptor agonist/antagonists (60, 61). In addition, they can also be used for imaging (62) and diagnostics (63), or as antimicrobial agent (64).
DARPins (Designed Ankyrin Repeat Proteins) may be especially interesting due to their high stability and high production levels when expressed in bacteria, their potential multivalence, possibility of site-specific conjugations, and their potential picomolar affinities (65). They are a cost-effective solution for large-scale production (the costs of generating a new DARPin are similar to making a custom monoclonal antibody), for detection or interference inside targeted cells, and for multiplex applications. They have been successfully used for chimeric antigen receptor-T cells, for viral manipulations and for cytoplasmic markers. Like for many scaffolds, the research market is not yet served commercially, but only through academic collaborations. Because of the attractive profit margins, commercial use has focused on therapeutics (66).
Fynomers are binding proteins derived from the SH3 domain of human tyrosine kinase Fyn. These scaffold proteins are composed of two β-sheets and two exposed loops for binding to target proteins. The Fyn SH3 domain is a particularly attractive scaffold for the generation of binding proteins because the resulting Fynomer (67):
- Is expressed in bacteria in soluble form in high amounts
- Is monomeric and does not aggregate when stored in solution
- Is very stable (Tm ~70°C)
- Lacks cysteine residues
- Is of human origin featuring an amino acid sequence completely conserved from mouse to man and, hence, is considered to be non-immunogenic.
- Fynomer candidates can be panned from a phage display library
- High-affinity Fynomers can be obtained after randomization of the RT- or n-src-loop, and subsequent affinity maturation selection
Fynomers can be fused to N-terminal and/or C-terminal ends of antibodies to generate bispecific therapeutics (FynomAbs). For example, the bispecific HER2/CD3 targeting FynomAbs redirect T cells to HER2-expressing tumour cells, causing tumour cell lysis in multiple cell lines, using only picomolar quantities. The activity was found to be highly specific, as no lysis of cells was observed in the absence of HER2 expression (68). FynomAb COVA322 was designed to simultaneously inhibit the activity of both TNF and IL-17A for treatment of rheumatoid arthritis (69). COVA322 is currently being evaluated in a phase II clinical trial (NCT02243787).
Kunitz domains are ~60-amino-acid peptides derived from the active motif of Kunitz-type protease inhibitors such as Aprotinin (bovine pancreatic trypsin inhibitor), Alzheimer’s amyloid Precursor Protein, and Tissue Factor Pathway Inhibitor. Most important for library generation, they possess loop regions that can be mutated without destabilizing the structural framework. Thus, phage display libraries can be panned for the desired binding capacity. These binders have been used primarily for clinical applications (70, 71).
Aptamers
Aptamers are oligonucleotides, such as ribonucleic acid (RNA) and single-strand deoxyribonucleic acid (ssDNA), chemically modified to protect their molecular integrity in physiological environment. Generally, aptamers fold into a specific tertiary structure and bind to targets such as proteins, metals, and molecules. The structural folding of oligonucleotides provides high affinity and excellent specificity toward targets, thus making them analogues of antibodies. Aptamers are generated by an in vitro molecular evolution method known as Systematic Evolution of Ligands by EXponential enrichment (SELEX) (72). The power of SELEX comes from the extremely large libraries of randomized nucleic acid sequences that can be efficiently screened for molecules with functional properties defined by the selection conditions. Such screening can be conducted against a variety of target molecules or elements, such as small compounds, proteins, nanoparticles, or live cells. Hence, aptamers can be used as reagents for affinity purification as well as detection (biosensors) and therapeutics to replace antibodies.
Stability of aptamers
Oligonucleotides are intrinsically unstable due to nucleolytic degradation by intracellular and extracellular nucleases. Such degradation is evaded by chemical modifications while the plasticity of the aptamer remains unaffected. Several options of modification are available and used to stabilise the aptamer. In addition to stability, it also turned out that finding high affinity aptamers against “difficult” human proteins were easier to find after modification (74).
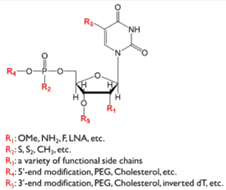
Figure 7. Nucleotide with the options for chemical modification to stabilise the oligonucleotide aptamer. (Figure 2 from ref 73)
Aptamers for detection
The company SomaLogic developed a proteomic technology for biomarker discovery that is capable of simultaneously measuring more than a thousand proteins from small volumes of biological samples such as plasma, tissues, or cells. The technology is based on oligonucleotides with chemical modifications that greatly increase the physicochemical diversity of nucleic acid-based ligands. Such modifications introduce functional groups that are absent in natural nucleic acids but are often found in protein–protein, small molecule–protein, and antibody–antigen interactions. This new class of reagents are called SOMAmers (Slow Off-rate Modified Aptamers). The use of these modifications expands the range of possible targets for SELEX, results in improved binding properties, and facilitates selection of SOMAmers with slow dissociation rates (74). These slow off-rates in combination with the ability to hybridize to complementary sequences on a nucleic acid array have led to the development of the so-called SOMAscan.
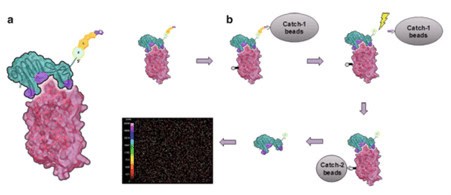
Figure 8. (a) Schematic representation of an interaction between a SOMAmer (green) and its protein target (red). Modified residues at the interface are shown in purple. For the assay, the SOMAmer contains a biotin (B), a photocleavable linker (L), and a fluorophore (F) at the 5’end.
(b) Schematic sequence of assay steps leading to quantitative readout of target proteins (clockwise from top left): SOMAmer–protein binding, capture onto streptavidin-coated beads (“catch-1”), biotin-labeling of proteins, photocleavage followed by kinetic challenge, second biotin capture (“catch-2”), release from complexes into solution at high pH, and hybridization to complementary aptamer microarray. SOMAmers are detected by fluorescent tags when the array is scanned. (Figure 20.2 from ref 74)
This multiplex system benefits from high specificity due to a stringent wash (kinetic challenge, fig 8) with a polyanionic competitor that selectively disrupts non-cognate complexes. Because dissociation rates of cognate SOMAmer–protein interactions are much slower than those of nonspecific interactions. The quantity of each analyte is measured by the intensity of fluorescence on the microarray. Despite its complexity, the assay has a large overall dynamic range of 7 logs (from 100 fM to 1mM), low limits of detection (0.3 pM median), and excellent reproducibility (5% median intra-run and inter-run coefficient of variance).
This SOMAmer platform has been used successfully for biomarker discovery in a large set of diseases, and a detailed review was published recently (75).
Aptamers for therapeutics
Aptamers could be used as carriers of therapy for accurate delivery of therapeutic drugs, such as chemotherapy drugs, small interfering RNAs, microRNAs, antisense oligos, or toxins, to the target cells but not to normal cells (76). These aptamer-drug conjugates (ApDCs) have gradually become effective drug delivery systems and attracted widespread attention (77). The simplest form of aptamer drug conjugation is non-covalent. Earlier aptamer-chemotherapy drug conjugates were constructed by non-covalent conjugation. Based on the various chemical properties of aptamers, many studies have proposed different aptamer-drug conjugation methods, such as covalent crosslinking, biocompatible spacers, cleavable linkers, and hydrophobic interaction. Examples are summarised in a new review (78).
Aptamer-mediated nucleic acid delivery for therapeutics is currently intensely tried and developed. Examples of siRNA delivery with reported success are: multivalent aptamer-siRNA conjugates (79), aptamer-based liposomes (80), bivalent aptamer-siRNA chimera (81) and multifunctional origami-based nanocarrier for codelivery of drugs and anti-sense oligonucleotides (128, 82).
Bispecific Aptamers have been developed, inspired by bispecific antibody therapies. Boltz et al. developed a stable nucleotide-based bispecific aptamer that mediates the lysis of c-Met-positive tumours, thus opening up new avenues for tumour therapy (83). In particular applications involving Natural Killer Cells have been developed (78).
Aptamers can be developed into cell growth factor mimics (84, 85) or extracellular matrix mimics (86, 87, 88), which can provide a favourable growth environment for cells, or simulate the related mechanisms in vivo, thus contributing to cell signal transduction and growth, as well as tissue repair.
The specificity and affinity of aptamers make them fit as mediators in cell recruitment for tissue repair or tumour targeting. Most repair studies focus on bone and cartilage tissue repair, in which the aptamers specific for Bone Marrow-derived Mesenchymal Stem Cells (BM-MSCs) are loaded onto the scaffold in bone therapy, and BM-MSCs are recruited to achieve regeneration therapy (89). However, BM-MSCs are also key contributors to the tumour microenvironment (TME), and participate in tumour progression and promoting tumour metastasis by establishing a favourable TME (90). Hence, recruitment of BM-MSCs are prevented as an aptamer-mediated therapy through intervention of the platelet-derived growth factor receptor β (PDGFRβ). Camorani et al. used anti-PDGFRβ aptamer (Gint4.T) as a novel therapeutic tool to inhibit the homing of BM-MSCs to the triple-negative breast cancer (TNBC) microenvironment and TNBC-stromal interactions, ultimately interfering with BM-MSC-dependent pro-invasive activity (91).
To date, eleven aptamers have been clinically translated and are in various stages of clinical studies. The therapeutic scope of these aptamers includes eye disease, coagulation, oncology, and inflammation (reviewed in 78).
References
- Baker M. When antibodies mislead: the quest for validation. Nature. 2020 Sep;585(7824):313-314. doi: 10.1038/d41586-020-02549-1. PMID: 32895531.
- Voskuil JLA, Bandrowski A, Begley CG, Bradbury ARM, Chalmers AD, Gomes AV, Hardcastle T, Lund-Johansen F, Plückthun A, Roncador G, Solache A, Taussig MJ, Trimmer JS, Williams C, Goodman SL. The Antibody Society’s antibody validation webinar series. MAbs. 2020 Jan-Dec;12(1):1794421. doi: 10.1080/19420862.2020.1794421. PMID: 32748696; PMCID: PMC7531563.
- Voskuil JLA. The challenges with the validation of research antibodies. F1000Res. 2017 Feb 17;6:161. doi: 10.12688/f1000research.10851.1. PMID: 28357047; PMCID: PMC5333605.
- Miersch S, Sidhu SS. Synthetic antibodies: concepts, potential and practical considerations. Methods. 2012 Aug;57(4):486-98. doi: 10.1016/j.ymeth.2012.06.012. Epub 2012 Jun 27. PMID: 22750306
- Vazquez-Lombardi R, Phan TG, Zimmermann C, Lowe D, Jermutus L, Christ D. Challenges and opportunities for non-antibody scaffold drugs. Drug Discov Today. 2015 Oct;20(10):1271-83. doi: 10.1016/j.drudis.2015.09.004. PMID: 26360055.
- Binz HK, Plückthun A. Engineered proteins as specific binding reagents. Curr Opin Biotechnol. 2005 Aug;16(4):459-69. doi: 10.1016/j.copbio.2005.06.005. PMID: 16005204.
- Frejd FY, Kim KT. Affibody molecules as engineered protein drugs. Exp Mol Med. 2017 Mar 24;49(3):e306. doi: 10.1038/emm.2017.35. PMID: 28336959.
- Scott CJ, Taggart CC. Biologic protease inhibitors as novel therapeutic agents. Biochimie. 2010 Nov;92(11):1681-8. doi: 10.1016/j.biochi.2010.03.010. PMID: 20346385.
- Heinzelman P, Krais J, Ruben E, Pantazes R. Engineering pH responsive fibronectin domains for biomedical applications. J Biol Eng. 2015 May 15;9:6. doi: 10.1186/s13036-015-0004-1. PMID: 26106447
- Owens B. Faster, deeper, smaller-the rise of antibody-like scaffolds. Nat Biotechnol. 2017 Jul 12;35(7):602-603. doi: 10.1038/nbt0717-602. PMID: 28700554.
- Koide A, Bailey CW, Huang X, Koide S. The fibronectin type III domain as a scaffold for novel binding proteins. J Mol Biol. 1998 Dec 11;284(4):1141-51. doi: 10.1006/jmbi.1998.2238. PMID: 9837732.
- Koide A, Koide S. Monobodies: antibody mimics based on the scaffold of the fibronectin type III domain. Methods Mol Biol. 2007;352:95-109. doi: 10.1385/1-59745-187-8:95. PMID: 17041261.
- Fellouse FA, Li B, Compaan DM, Peden AA, Hymowitz SG, Sidhu SS. Molecular recognition by a binary code. J Mol Biol. 2005 May 20;348(5):1153-62. doi: 10.1016/j.jmb.2005.03.041. PMID: 15854651.
- Koide A, Gilbreth RN, Esaki K, Tereshko V, Koide S. High-affinity single-domain binding proteins with a binary-code interface. Proc Natl Acad Sci U S A. 2007 Apr 17;104(16):6632-7. doi: 10.1073/pnas.0700149104. PMID: 17420456.
- Hackel BJ, Wittrup KD. The full amino acid repertoire is superior to serine/tyrosine for selection of high affinity immunoglobulin G binders from the fibronectin scaffold. Protein Eng Des Sel. 2010 Apr;23(4):211-9. doi: 10.1093/protein/gzp083. PMID: 20067921.
- Hantschel O, Biancalana M, Koide S. Monobodies as enabling tools for structural and mechanistic biology. Curr Opin Struct Biol. 2020 Feb;60:167-174. doi: 10.1016/j.sbi.2020.01.015. PMID: 32145686
- Lipovsek D. Adnectins: engineered target-binding protein therapeutics. Protein Eng Des Sel. 2011 Jan;24(1-2):3-9. doi: 10.1093/protein/gzq097. PMID: 21068165.
- Chandler and Buckle 2020, Chandler PG, Buckle AM. Development and Differentiation in Monobodies Based on the Fibronectin Type 3 Domain. Cells. 2020 Mar 4;9(3):610. doi: 10.3390/cells9030610. PMID: 32143310
- Prabakaran P, Vu BK, Gan J, Feng Y, Dimitrov DS, Ji X. Structure of an isolated unglycosylated antibody C(H)2 domain. Acta Crystallogr D Biol Crystallogr. 2008 Oct;64(Pt 10):1062-7. doi: 10.1107/S0907444908025274. PMID: 18931413.
- Ullman C, Mathonet P, Oleksy A, Diamandakis A, Tomei L, Demartis A, Nardi C, Sambucini S, Missineo A, Alt K, Hagemeyer CE, Harris M, Hedt A, Weis R, Gehlsen KR. High Affinity Binders to EphA2 Isolated from Abdurin Scaffold Libraries; Characterization, Binding and Tumor Targeting. PLoS One. 2015 Aug 27;10(8):e0135278. doi: 10.1371/journal.pone.0135278. PMID: 26313909.
- Deonarain MP, Xue Q. Tackling solid tumour therapy with small-format drug conjugates. Antib Ther. 2020 Nov 25;3(4):237-245. doi: 10.1093/abt/tbaa024. PMID: 33928231; PMCID: PMC7990258.
- Tashiro M, Tejero R, Zimmerman DE, Celda B, Nilsson B, Montelione GT. High-resolution solution NMR structure of the Z domain of staphylococcal protein A. J Mol Biol. 1997 Oct 3;272(4):573-90. doi: 10.1006/jmbi.1997.1265. PMID: 9325113.
- Nord K, Gunneriusson E, Ringdahl J, Ståhl S, Uhlén M, Nygren PA. Binding proteins selected from combinatorial libraries of an alpha-helical bacterial receptor domain. Nat Biotechnol. 1997 Aug;15(8):772-7. doi: 10.1038/nbt0897-772. PMID: 9255793.
- Frejd FY, Kim KT. Affibody molecules as engineered protein drugs. Exp Mol Med. 2017 Mar 24;49(3):e306. doi: 10.1038/emm.2017.35. PMID: 28336959; PMCID: PMC5382565.
- Tolmachev V, Orlova A. Affibody Molecules as Targeting Vectors for PET Imaging. Cancers (Basel). 2020 Mar 11;12(3):651. doi: 10.3390/cancers12030651. PMID: 32168760
- Garousi J, Andersson KG, Dam JH, Olsen BB, Mitran B, Orlova A, Buijs J, Ståhl S, Löfblom J, Thisgaard H, Tolmachev V. The use of radiocobalt as a label improves imaging of EGFR using DOTA-conjugated Affibody molecule. Sci Rep. 2017 Jul 20;7(1):5961. doi: 10.1038/s41598-017-05700-7. PMID: 28729680.
- LaFleur DW, Abramyan D, Kanakaraj P, Smith RG, Shah RR, Wang G, Yao XT, Kankanala S, Boyd E, Zaritskaya L, Nam V, Puffer BA, Buasen P, Kaithamana S, Burnette AF, Krishnamurthy R, Patel D, Roschke VV, Kiener PA, Hilbert DM, Barbas CF 3rd. Monoclonal antibody therapeutics with up to five specificities: functional enhancement through fusion of target-specific peptides. MAbs. 2013 Mar-Apr;5(2):208-18. doi: 10.4161/mabs.23043. PMID: 23575268
- Yu F, Gudmundsdotter L, Akal A, Gunneriusson E, Frejd F, Nygren PÅ. An affibody-adalimumab hybrid blocks combined IL-6 and TNF-triggered serum amyloid A secretion in vivo. MAbs. 2014;6(6):1598-607. doi: 10.4161/mabs.36089. PMID: 25484067
- Schardt JS, Oubaid JM, Williams SC, Howard JL, Aloimonos CM, Bookstaver ML, Lamichhane TN, Sokic S, Liyasova MS, O’Neill M, Andresson T, Hussain A, Lipkowitz S, Jay SM. Engineered Multivalency Enhances Affibody-Based HER3 Inhibition and Downregulation in Cancer Cells. Mol Pharm. 2017 Apr 3;14(4):1047-1056. doi: 10.1021/acs.molpharmaceut.6b00919. Epub 2017 Mar 8. PMID: 28248115.
- Mączyńska J, Da Pieve C, Burley TA, Raes F, Shah A, Saczko J, Harrington KJ, Kramer-Marek G. Immunomodulatory activity of IR700-labelled affibody targeting HER2. Cell Death Dis. 2020 Oct 20;11(10):886. doi: 10.1038/s41419-020-03077-6. PMID: 33082328.
- Zhu J, Kamara S, Cen D, Tang W, Gu M, Ci X, Chen J, Wang L, Zhu S, Jiang P, Chen S, Xue X, Zhang L. Generation of novel affibody molecules targeting the EBV LMP2A N-terminal domain with inhibiting effects on the proliferation of nasopharyngeal carcinoma cells. Cell Death Dis. 2020 Apr 1;11(4):213. doi: 10.1038/s41419-020-2410-7. Erratum in: Cell Death Dis. 2020 Jun 30;11(6):494. PMID: 32238802.
- Tiede C, Tang AA, Deacon SE, Mandal U, Nettleship JE, Owen RL, George SE, Harrison DJ, Owens RJ, Tomlinson DC, McPherson MJ. Adhiron: a stable and versatile peptide display scaffold for molecular recognition applications. Protein Eng Des Sel. 2014 May;27(5):145-55. doi: 10.1093/protein/gzu007. PMID: 24668773.
- Stadler LK, Hoffmann T, Tomlinson DC, Song Q, Lee T, Busby M, Nyathi Y, Gendra E, Tiede C, Flanagan K, Cockell SJ, Wipat A, Harwood C, Wagner SD, Knowles MA, Davis JJ, Keegan N, Ferrigno PK. Structure-function studies of an engineered scaffold protein derived from Stefin A. II: Development and applications of the SQT variant. Protein Eng Des Sel. 2011 Sep;24(9):751-63. doi: 10.1093/protein/gzr019. Epub 2011 May 25. PMID: 21616931.
- Tiede C, Bedford R, Heseltine SJ, Smith G, Wijetunga I, Ross R, AlQallaf D, Roberts AP, Balls A, Curd A, Hughes RE, Martin H, Needham SR, Zanetti-Domingues LC, Sadigh Y, Peacock TP, Tang AA, Gibson N, Kyle H, Platt GW, Ingram N, Taylor T, Coletta LP, Manfield I, Knowles M, Bell S, Esteves F, Maqbool A, Prasad RK, Drinkhill M, Bon RS, Patel V, Goodchild SA, Martin-Fernandez M, Owens RJ, Nettleship JE, Webb ME, Harrison M, Lippiat JD, Ponnambalam S, Peckham M, Smith A, Ferrigno PK, Johnson M, McPherson MJ, Tomlinson DC. Affimer proteins are versatile and renewable affinity reagents. Elife. 2017 Jun 27;6:e24903. doi: 10.7554/eLife.24903. PMID: 28654419
- Tans R, van Rijswijck DMH, Davidson A, Hannam R, Ricketts B, Tack CJ, Wessels HJCT, Gloerich J, van Gool AJ. Affimers as an alternative to antibodies for protein biomarker enrichment. Protein Expr Purif. 2020 Oct;174:105677. doi: 10.1016/j.pep.2020.105677. PMID: 32461183.
- Klont F, Hadderingh M, Horvatovich P, Ten Hacken NHT, Bischoff R. Affimers as an Alternative to Antibodies in an Affinity LC-MS Assay for Quantification of the Soluble Receptor of Advanced Glycation End-Products (sRAGE) in Human Serum. J Proteome Res. 2018 Aug 3;17(8):2892-2899. doi: 10.1021/acs.jproteome.8b00414. PMID: 30005571.
- Adamson H, Nicholl A, Tiede C, Tang AA, Davidson A, Curd H, Wignall A, Ford R, Nuttall J, McPherson MJ, Johnson M, Tomlinson DC. Affimers as anti-idiotypic affinity reagents for pharmacokinetic analysis of biotherapeutics. Biotechniques. 2019 Dec;67(6):261-269. doi: 10.2144/btn-2019-0089. PMID: 31823668.
- Mouratou B, Schaeffer F, Guilvout I, Tello-Manigne D, Pugsley AP, Alzari PM, Pecorari F. Remodeling a DNA-binding protein as a specific in vivo inhibitor of bacterial secretin PulD. Proc Natl Acad Sci U S A. 2007 Nov 13;104(46):17983-8. doi: 10.1073/pnas.0702963104. PMID: 17984049.
- Béhar G, Bellinzoni M, Maillasson M, Paillard-Laurance L, Alzari PM, He X, Mouratou B, Pecorari F. Tolerance of the archaeal Sac7d scaffold protein to alternative library designs: characterization of anti-immunoglobulin G Affitins. Protein Eng Des Sel. 2013 Apr;26(4):267-75. doi: 10.1093/protein/gzs106. PMID: 23315487.
- Béhar G, Renodon-Cornière A, Mouratou B, Pecorari F. Affitins as robust tailored reagents for affinity chromatography purification of antibodies and non-immunoglobulin proteins. J Chromatogr A. 2016 Apr 8;1441:44-51. doi: 10.1016/j.chroma.2016.02.068. PMID: 26952369.
- Béhar G, Renodon-Cornière A, Kambarev S, Vukojicic P, Caroff N, Corvec S, Mouratou B, Pecorari F. Whole-bacterium ribosome display selection for isolation of highly specific anti-Staphyloccocus aureus Affitins for detection- and capture-based biomedical applications. Biotechnol Bioeng. 2019 Aug;116(8):1844-1855. doi: 10.1002/bit.26989. Epub 2019 May 15. PMID: 30982947.
- Kalichuk V, Kambarev S, Béhar G, Chalopin B, Renodon-Cornière A, Mouratou B, Pecorari F. Affitins: Ribosome Display for Selection of Aho7c-Based Affinity Proteins. Methods Mol Biol. 2020;2070:19-41. doi: 10.1007/978-1-4939-9853-1_2. PMID: 31625088.
- Gera N, Hussain M, Wright RC, Rao BM. Highly stable binding proteins derived from the hyperthermophilic Sso7d scaffold. J Mol Biol. 2011 Jun 17;409(4):601-16. doi: 10.1016/j.jmb.2011.04.020. PMID: 21515282.
- Beste G, Schmidt FS, Stibora T, Skerra A. Small antibody-like proteins with prescribed ligand specificities derived from the lipocalin fold. Proc Natl Acad Sci U S A. 1999 Mar 2;96(5):1898-903. doi: 10.1073/pnas.96.5.1898. PMID: 10051566
- Schlehuber S, Beste G, Skerra A. A novel type of receptor protein, based on the lipocalin scaffold, with specificity for digoxigenin. J Mol Biol. 2000 Apr 14;297(5):1105-20. doi: 10.1006/jmbi.2000.3646. PMID: 10764576.
- Schiefner A, Gebauer M, Richter A, Skerra A. Anticalins Reveal High Plasticity in the Mode of Complex Formation with a Common Tumor Antigen. Structure. 2018 Apr 3;26(4):649-656.e3. doi: 10.1016/j.str.2018.02.003. PMID: 29526433.
- Richter A, Skerra A. Anticalins directed against vascular endothelial growth factor receptor 3 (VEGFR-3) with picomolar affinities show potential for medical therapy and in vivo imaging. Biol Chem. 2017 Jan 1;398(1):39-55. doi: 10.1515/hsz-2016-0195. PMID: 27458663.
- Gille H, Hülsmeyer M, Trentmann S, Matschiner G, Christian HJ, Meyer T, Amirkhosravi A, Audoly LP, Hohlbaum AM, Skerra A. Functional characterization of a VEGF-A-targeting Anticalin, prototype of a novel therapeutic human protein class. Angiogenesis. 2016 Jan;19(1):79-94. doi: 10.1007/s10456-015-9490-5. PMID: 26650228.
- Deuschle FC, Ilyukhina E, Skerra A. Anticalin® proteins: from bench to bedside. Expert Opin Biol Ther. 2021 Apr;21(4):509-518. doi: 10.1080/14712598.2021.1839046. PMID: 33074019.
- Li H, Liu L, Ning B, Sun Z, Yao S, Jiang Y, Liu J. Selection of an artificial paraquat-specific binding protein from a ribosome display library based on a lipocalin scaffold. Biotechnol Appl Biochem. 2020 Nov 10. doi: 10.1002/bab.2059. Epub ahead of print. PMID: 33169873.
- Morita H, Shimbo K, Shigemori H, Kobayashi J. Antimitotic activity of moroidin, a bicyclic peptide from the seeds of Celosia argentea. Bioorg Med Chem Lett. 2000 Mar 6;10(5):469-71. doi: 10.1016/s0960-894x(00)00029-9. PMID: 10743950.
- Capani F, Deerinck TJ, Ellisman MH, Bushong E, Bobik M, Martone ME. Phalloidin-eosin followed by photo-oxidation: a novel method for localizing F-actin at the light and electron microscopic levels. J Histochem Cytochem. 2001 Nov;49(11):1351-61. doi: 10.1177/002215540104901103. PMID: 11668188.
- Lindell TJ, Weinberg F, Morris PW, Roeder RG, Rutter WJ. Specific inhibition of nuclear RNA polymerase II by alpha-amanitin. Science. 1970 Oct 23;170(3956):447-9. doi: 10.1126/science.170.3956.447. PMID: 4918258.
- Ahangarzadeh S, Kanafi MM, Hosseinzadeh S, Mokhtarzadeh A, Barati M, Ranjbari J, Tayebi L. Bicyclic peptides: types, synthesis and applications. Drug Discov Today. 2019 Jun;24(6):1311-1319. doi: 10.1016/j.drudis.2019.05.008. PMID: 31102732.
- Roodbeen R, Paaske B, Jiang L, Jensen JK, Christensen A, Nielsen JT, Huang M, Mulder FA, Nielsen NC, Andreasen PA, Jensen KJ. Bicyclic peptide inhibitor of urokinase-type plasminogen activator: mode of action. Chembiochem. 2013 Nov 4;14(16):2179-88. doi: 10.1002/cbic.201300335. PMID: 24115455.
- Islam NM, Kato T, Nishino N, Kim HJ, Ito A, Yoshida M. Bicyclic peptides as potent inhibitors of histone deacetylases: optimization of alkyl loop length. Bioorg Med Chem Lett. 2010 Feb 1;20(3):997-9. doi: 10.1016/j.bmcl.2009.12.054. PMID: 20045316.
- Teufel DP, Bennett G, Harrison H, van Rietschoten K, Pavan S, Stace C, Le Floch F, Van Bergen T, Vermassen E, Barbeaux P, Hu TT, Feyen JHM, Vanhove M. Stable and Long-Lasting, Novel Bicyclic Peptide Plasma Kallikrein Inhibitors for the Treatment of Diabetic Macular Edema. J Med Chem. 2018 Apr 12;61(7):2823-2836. doi: 10.1021/acs.jmedchem.7b01625. PMID: 29517911.
- Lian W, Upadhyaya P, Rhodes CA, Liu Y, Pei D. Screening bicyclic peptide libraries for protein-protein interaction inhibitors: discovery of a tumor necrosis factor-α antagonist. J Am Chem Soc. 2013 Aug 14;135(32):11990-5. doi: 10.1021/ja405106u. PMID: 23865589
- Guardiola S, Seco J, Varese M, Díaz-Lobo M, García J, Teixidó M, Nevola L, Giralt E. Toward a Novel Drug To Target the EGF-EGFR Interaction: Design of Metabolically Stable Bicyclic Peptides. Chembiochem. 2018 Jan 4;19(1):76-84. doi: 10.1002/cbic.201700519. PMID: 29105934.
- Potterat O, Wagner K, Gemmecker G, Mack J, Puder C, Vettermann R, Streicher R. BI-32169, a bicyclic 19-peptide with strong glucagon receptor antagonist activity from Streptomyces sp. J Nat Prod. 2004 Sep;67(9):1528-31. doi: 10.1021/np040093o. PMID: 15387654
- Jia H, Bagherzadeh A, Hartzoulakis B, Jarvis A, Löhr M, Shaikh S, Aqil R, Cheng L, Tickner M, Esposito D, Harris R, Driscoll PC, Selwood DL, Zachary IC. Characterization of a bicyclic peptide neuropilin-1 (NP-1) antagonist (EG3287) reveals importance of vascular endothelial growth factor exon 8 for NP-1 binding and role of NP-1 in KDR signaling. J Biol Chem. 2006 May 12;281(19):13493-13502. doi: 10.1074/jbc.M512121200. PMID: 16513643.
- Park JA, Lee YJ, Lee JW, Lee KC, An GI, Kim KM, Kim BI, Kim TJ, Kim JY. Cyclic RGD Peptides Incorporating Cycloalkanes: Synthesis and Evaluation as PET Radiotracers for Tumor Imaging. ACS Med Chem Lett. 2014 Jul 10;5(9):979-82. doi: 10.1021/ml500135t. PMID: 25221652
- Eder M, Pavan S, Bauder-Wüst U, van Rietschoten K, Baranski AC, Harrison H, Campbell S, Stace CL, Walker EH, Chen L, Bennett G, Mudd G, Schierbaum U, Leotta K, Haberkorn U, Kopka K, Teufel DP. Bicyclic Peptides as a New Modality for Imaging and Targeting of Proteins Overexpressed by Tumors. Cancer Res. 2019 Feb 15;79(4):841-852. doi: 10.1158/0008-5472.CAN-18-0238. PMID: 30606721.
- Di Bonaventura I, Jin X, Visini R, Probst D, Javor S, Gan BH, Michaud G, Natalello A, Doglia SM, Köhler T, van Delden C, Stocker A, Darbre T, Reymond JL. Chemical space guided discovery of antimicrobial bridged bicyclic peptides against Pseudomonas aeruginosa and its biofilms. Chem Sci. 2017 Oct 1;8(10):6784-6798. doi: 10.1039/c7sc01314k. Epub 2017 Jul 13. PMID: 29147502
- Plückthun A. Designed ankyrin repeat proteins (DARPins): binding proteins for research, diagnostics, and therapy. Annu Rev Pharmacol Toxicol. 2015;55:489-511. doi: 10.1146/annurev-pharmtox-010611-134654. PMID: 25562645.
- Kumar A, Balbach J. Folding and Stability of Ankyrin Repeats Control Biological Protein Function. Biomolecules. 2021 Jun 5;11(6):840. doi: 10.3390/biom11060840. PMID: 34198779; PMCID: PMC8229355.
- Schlatter D, Brack S, Banner DW, Batey S, Benz J, Bertschinger J, Huber W, Joseph C, Rufer A, van der Klooster A, Weber M, Grabulovski D, Hennig M. Generation, characterization and structural data of chymase binding proteins based on the human Fyn kinase SH3 domain. MAbs. 2012 Jul-Aug;4(4):497-508. doi: 10.4161/mabs.20452. PMID: 22653218.
- R. Woods, S. Brack, K. Klupsch, U. Wuellner, F. Buller, R. Santimaria. Fynomer-antibody fusions targeting HER2 and CD3 for selective killing of HER2 overexpressing tumor cells. J. Clin. Oncol., 32 (2014); 3066-3066, 10.1200/jco.2014.32.15_suppl.3066
- Silacci M, Lembke W, Woods R, Attinger-Toller I, Baenziger-Tobler N, Batey S, Santimaria R, von der Bey U, Koenig-Friedrich S, Zha W, Schlereth B, Locher M, Bertschinger J, Grabulovski D. Discovery and characterization of COVA322, a clinical-stage bispecific TNF/IL-17A inhibitor for the treatment of inflammatory diseases. MAbs. 2016;8(1):141-9. doi: 10.1080/19420862.2015.1093266. PMID: 26390837.
- Hosse RJ, Rothe A, Power BE. A new generation of protein display scaffolds for molecular recognition. Protein Sci. 2006 Jan;15(1):14-27. doi: 10.1110/ps.051817606. PMID: 16373474.
- Simeon R, Chen Z. In vitro-engineered non-antibody protein therapeutics. Protein Cell. 2018 Jan;9(1):3-14. doi: 10.1007/s13238-017-0386-6. PMID: 28271446.
- Nakamura Y, Ishiguro A, Miyakawa S. RNA plasticity and selectivity applicable to therapeutics and novel biosensor development. Genes Cells. 2012 May;17(5):344-64. doi: 10.1111/j.1365-2443.2012.01596.x. PMID: 22487172.
- Adachi T, Nakamura Y. Aptamers: A Review of Their Chemical Properties and Modifications for Therapeutic Application. Molecules. 2019 Nov 21;24(23):4229. doi: 10.3390/molecules24234229. PMID: 31766318; PMCID: PMC6930564.
- Mehan MR, Ostroff R, Wilcox SK, Steele F, Schneider D, Jarvis TC, Baird GS, Gold L, Janjic N. Highly Multiplexed Proteomic Platform for Biomarker Discovery, Diagnostics, and Therapeutics. In: Lambris, J., Holers, V., Ricklin, D. (eds) Complement Therapeutics pp 283-300. Advances in Experimental Medicine and Biology, vol 735. Springer, New York, NY. https://doi.org/10.1007/978-1-4614-4118-2_20
- Huang J, Chen X, Fu X, Li Z, Huang Y, Liang C. Advances in Aptamer-Based Biomarker Discovery. Front Cell Dev Biol. 2021 Mar 16;9:659760. doi: 10.3389/fcell.2021.659760. PMID: 33796540.
- Soldevilla MM, Meraviglia-Crivelli de Caso D, Menon AP, Pastor F. Aptamer-iRNAs as Therapeutics for Cancer Treatment. Pharmaceuticals (Basel). 2018 Oct 18;11(4):108. doi: 10.3390/ph11040108. PMID: 30340426.
- Xuan W, Peng Y, Deng Z, Peng T, Kuai H, Li Y, He J, Jin C, Liu Y, Wang R, Tan W. A basic insight into aptamer-drug conjugates (ApDCs). Biomaterials. 2018 Nov;182:216-226. doi: 10.1016/j.biomaterials.2018.08.021. Epub 2018 Aug 8. PMID: 30138784.
- Yuhan J, Zhu L, Zhu L, Huang K, He X, Xu W. Cell-specific aptamers as potential drugs in therapeutic applications: A review of current progress. J Control Release. 2022 May 4;346:405-420. doi: 10.1016/j.jconrel.2022.04.039. PMID: 35489545.
- Jeong H, Lee SH, Hwang Y, Yoo H, Jung H, Kim SH, Mok H. Multivalent Aptamer-RNA Conjugates for Simple and Efficient Delivery of Doxorubicin/siRNA into Multidrug-Resistant Cells. Macromol Biosci. 2017 Apr;17(4). doi: 10.1002/mabi.201600343. PMID: 27863037.
- Li L, Hou J, Liu X, Guo Y, Wu Y, Zhang L, Yang Z. Nucleolin-targeting liposomes guided by aptamer AS1411 for the delivery of siRNA for the treatment of malignant melanomas. Biomaterials. 2014 Apr;35(12):3840-50. doi: 10.1016/j.biomaterials.2014.01.019. Epub 2014 Jan 31. PMID: 24486214.
- Xue L, Maihle NJ, Yu X, Tang SC, Liu HY. Synergistic Targeting HER2 and EGFR with Bivalent Aptamer-siRNA Chimera Efficiently Inhibits HER2-Positive Tumor Growth. Mol Pharm. 2018 Nov 5;15(11):4801-4813. doi: 10.1021/acs.molpharmaceut.8b00388. PMID: 30222359.
- Pan Q, Nie C, Hu Y, Yi J, Liu C, Zhang J, He M, He M, Chen T, Chu X. Aptamer-Functionalized DNA Origami for Targeted Codelivery of Antisense Oligonucleotides and Doxorubicin to Enhance Therapy in Drug-Resistant Cancer Cells. ACS Appl Mater Interfaces. 2020 Jan 8;12(1):400-409. doi: 10.1021/acsami.9b20707. PMID: 31815420.
- Boltz A, Piater B, Toleikis L, Guenther R, Kolmar H, Hock B. Bi-specific aptamers mediating tumor cell lysis. J Biol Chem. 2011 Jun 17;286(24):21896-905. doi: 10.1074/jbc.M111.238261. PMID: 21531729.
- Yoshitomi T, Hayashi M, Oguro T, Kimura K, Wayama F, Furusho H, Yoshimoto K. Binding and Structural Properties of DNA Aptamers with VEGF-A-Mimic Activity. Mol Ther Nucleic Acids. 2020 Mar 6;19:1145-1152. doi: 10.1016/j.omtn.2019.12.034. PMID: 32059340.
- Ueki R , Atsuta S , Ueki A , Hoshiyama J , Li J , Hayashi Y , Sando S . DNA aptamer assemblies as fibroblast growth factor mimics and their application in stem cell culture. Chem Commun (Camb). 2019 Feb 26;55(18):2672-2675. doi: 10.1039/c8cc08080a. PMID: 30746545.
- Galli C, Parisi L, Piergianni M, Smerieri A, Passeri G, Guizzardi S, Costa F, Lumetti S, Manfredi E, Macaluso GM. Improved scaffold biocompatibility through anti-Fibronectin aptamer functionalization. Acta Biomater. 2016 Sep 15;42:147-156. doi: 10.1016/j.actbio.2016.07.035. PMID: 27449338.
- Zhang X, Battig MR, Chen N, Gaddes ER, Duncan KL, Wang Y. Chimeric Aptamer-Gelatin Hydrogels as an Extracellular Matrix Mimic for Loading Cells and Growth Factors. Biomacromolecules. 2016 Mar 14;17(3):778-87. doi: 10.1021/acs.biomac.5b01511. PMID: 26791559.
- Stejskalová A, Oliva N, England FJ, Almquist BD. Biologically Inspired, Cell-Selective Release of Aptamer-Trapped Growth Factors by Traction Forces. Adv Mater. 2019 Feb;31(7):e1806380. doi: 10.1002/adma.201806380. PMID: 30614086.
- Wang M, Wu H, Li Q, Yang Y, Che F, Wang G, Zhang L. Novel Aptamer-Functionalized Nanoparticles Enhances Bone Defect Repair By Improving Stem Cell Recruitment. Int J Nanomedicine. 2019 Nov 6;14:8707-8724. doi: 10.2147/IJN.S223164. Erratum in: Int J Nanomedicine. 2020 Aug 14;15:6093-6094. PMID: 31806966.
- Bussard KM, Mutkus L, Stumpf K, Gomez-Manzano C, Marini FC. Tumor-associated stromal cells as key contributors to the tumor microenvironment. Breast Cancer Res. 2016 Aug 11;18(1):84. doi: 10.1186/s13058-016-0740-2. PMID: 27515302.
- Camorani S, Hill BS, Fontanella R, Greco A, Gramanzini M, Auletta L, Gargiulo S, Albanese S, Lucarelli E, Cerchia L, Zannetti A. Inhibition of Bone Marrow-Derived Mesenchymal Stem Cells Homing Towards Triple-Negative Breast Cancer Microenvironment Using an Anti-PDGFRβ Aptamer. Theranostics. 2017 Aug 22;7(14):3595-3607. doi: 10.7150/thno.18974. PMID: 28912898.